9.1 DNA Sequence (IN PYTHON PLEASE) OLEASE LOOK AT PICTURE FOR REFRENCE
DNA, the carrier of genetic information in living things, has been used in criminal justice for decades. But how, exactly, does DNA profiling work? Given a sequence of DNA, how can forensic investigators identify to whom it belongs?
Well, DNA is really just a sequence of molecules called nucleotides, arranged into a particular shape (a double helix). Each nucleotide of DNA contains one of four different bases: adenine (A), cytosine (C), guanine (G), or thymine (T). Every human cell has billions of these nucleotides arranged in sequence. Some portions of this sequence (i.e. genome) are the same, or at least very similar, across almost all humans, but other portions of the sequence have a higher genetic diversity and thus vary more across the population.
One place where DNA tends to have high genetic diversity is in Short Tandem Repeats (STRs). An STR is a short sequence of DNA bases that tends to be repeated back-to-back numerous times at specific locations in DNA. The number of times any particular STR repeats varies a lot among different people. In the DNA samples below, for example, Alice has the STR AGAT repeated four times in her DNA, while Bob has the same STR repeated five times.
Using multiple STRs, rather than just one, can improve the accuracy of DNA profiling. If the probability that two people have the same number of repeats for a single STR is 5%, and the analyst looks at 10 different STRs, then the probability that two DNA samples match purely by chance is about 1 in 1 quadrillion (assuming all STRs are independent of each other). So if two DNA samples match in the number of repeats for each of the STRs, the analyst can be pretty confident they came from the same person. CODIS, The FBI's DNA database, uses 20 different STRs as part of its DNA profiling process.
TASK Define a function named computeLCS that receives a long DNA sequence and a STR (like AGAT, AATG, or TATC). The function returns the number of times the STR is repeated back-to-back within the DNA sequence. This is a core function that can be used in DNA profiling as discussed above.
HAVING ALOT OF TROUBLE
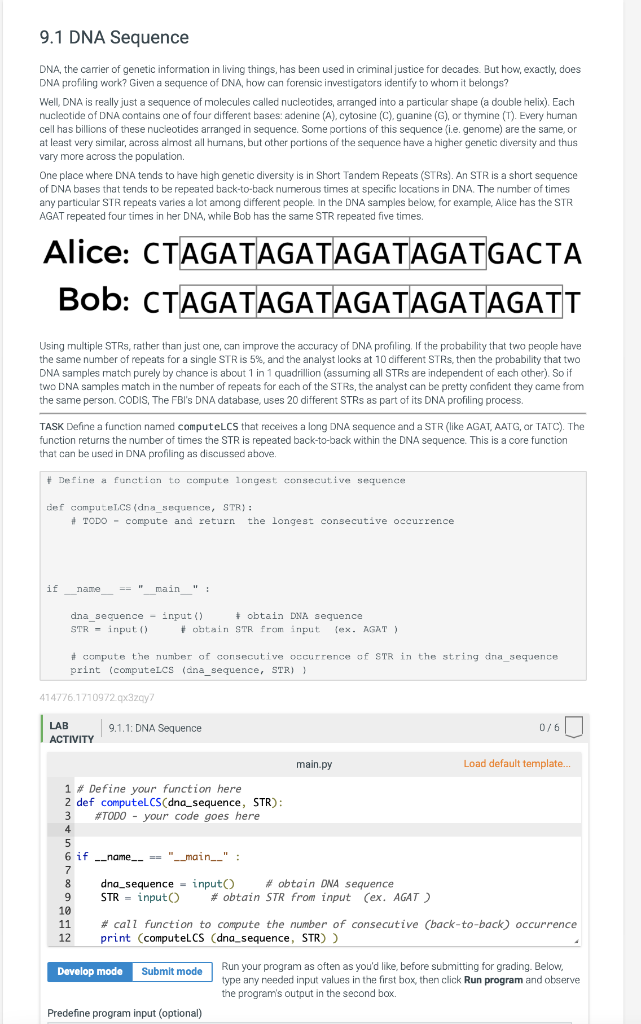
9.1 DNA Sequence DNA, the carrier of genetic information in living things, has been used in criminal justice for decades. But how, exactly, does DNA profiling work? Given a sequence of DNA, how can forensic investigators identify to whom it belongs? Well, DNA is really just a sequence of molecules called nucleotides, arranged into a particular shape (a double helix). Each nucleotide of DNA contains one of four different bases: adenine (A), cytosine (C), guanine (G), or thymine (T). Every human cell has billions of these nucleotides arranged in sequence. Some portions of this sequence (i.e. genome) are the same, or at least very similar, across almost all humans, but other portions of the sequence have a higher genetic diversity and thus vary more across the population. One place where DNA tends to have high genetic diversity is in Short Tandem Repeats (STRS). An STR is a short sequence of DNA bases that tends to be repeated back-to-back numerous times at specific locations in DNA. The number of times any particular STR repeats varies a lot among different people. In the DNA samples below, for example, Alice has the STR AGAT repeated four times in her DNA, while Bob has the same STR repeated five times. Alice: CTAGATAGATAGATAGATGACTA Bob: CTAGATAGATAGATAGATAGATT Using multiple STRS, rather than just one, can improve the accuracy of DNA profiling. If the probability that two people have the same number of repeats for a single STR is 5%, and the analyst looks at 10 different STRS, then the probability that two DNA samples match purely by chance is about 1 in 1 quadrillion (assuming all STRs are independent of each other). So if two DNA samples match in the number of repeats for each of the STRS, the analyst can be pretty confident they came from the same person. CODIS, The FBI's DNA database, uses 20 different STRs as part of its DNA profiling process. TASK Define a function named computeLCS that receives a long DNA sequence and a STR (like AGAT, AATG, or TATC). The function returns the number of times the STR is repeated back-to-back within the DNA sequence. This is a core function that can be used in DNA profiling as discussed above. #Define a function to compute longest consecutive sequence def computeLCS (dna_sequence, STR): # TODO - compute and return the longest consecutive occurrence if _name_____ == "____main__": dna_sequence input () STR input () #compute the number of consecutive occurrence of STR in the string dna_sequence print (computeLCS (dna_sequence, STR) ) 414776.1710972.qx3zqy7 LAB ACTIVITY - obtain DNA sequence #obtain STR from input (ex. AGAT) 9 1 # Define your function here 2 def computeLCS (dna_sequence, STR): 3 #TODO - your code goes here 4 10 11 9.1.1: DNA Sequence 5 6 if __name__ == "__main__": 7 8 dna_sequence=input() STR input() = Develop mode Submit mode main.py #obtain DNA sequence # obtain STR from input (ex. AGAT) Predefine program input (optional) 0/6 # call function to compute the number of consecutive (back-to-back) occurrence 12 print (computeLCS (dna_sequence, STR)) Load default template.. Run your program as often as you'd like, before submitting for grading. Below, type any needed input values in the first box, then click Run program and observe the program's output in the second box.
